plotMRCATest - Plot the results of a founder permutation test
Description¶
plotMRCATest plots the results of a founder permutation test performed with
testMRCA.
Usage¶
plotMRCATest(
data,
color = "black",
main_title = "MRCA Test",
style = c("histogram", "cdf"),
silent = FALSE,
...
)
Arguments¶
- data
- MRCATest object returned by testMRCA.
- color
- color of the histogram or lines.
- main_title
- string specifying the plot title.
- style
- type of plot to draw. One of:
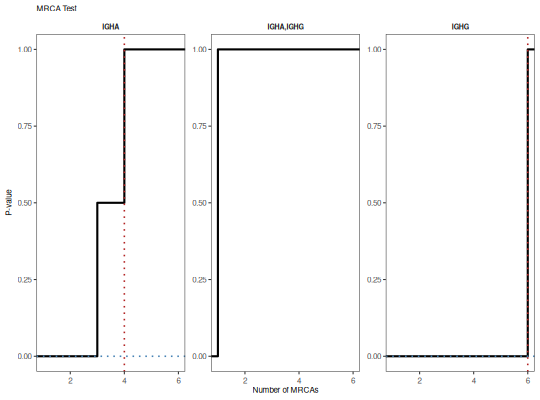
"histogram": histogram of the annotation count distribution with a red dotted line denoting the observed value."cdf": cumulative distribution function of annotation counts with a red dotted line denoting the observed value and a blue dotted line indicating the p-value.
- silent
- if
TRUEdo not draw the plot and just return the ggplot2 object; ifFALSEdraw the plot. - …
- additional arguments to pass to ggplot2::theme.
Value¶
A ggplot object defining the plot.
Examples¶
# Define example tree set
graphs <- ExampleTrees[1:10]
# Perform MRCA test on isotypes
x <- testMRCA(graphs, "c_call", nperm=10)
# Plot
plotMRCATest(x, color="steelblue", style="hist")
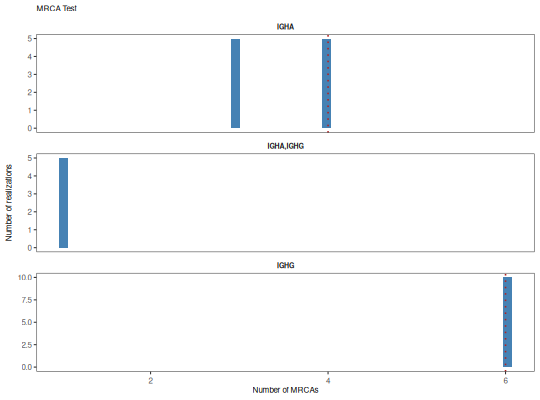
plotMRCATest(x, style="cdf")
See also¶
See testEdges for performing the test.